Revolutionary Intestinal Protein Analysis Technique Unveils Health Insights and Disease Targets
January 20, 2025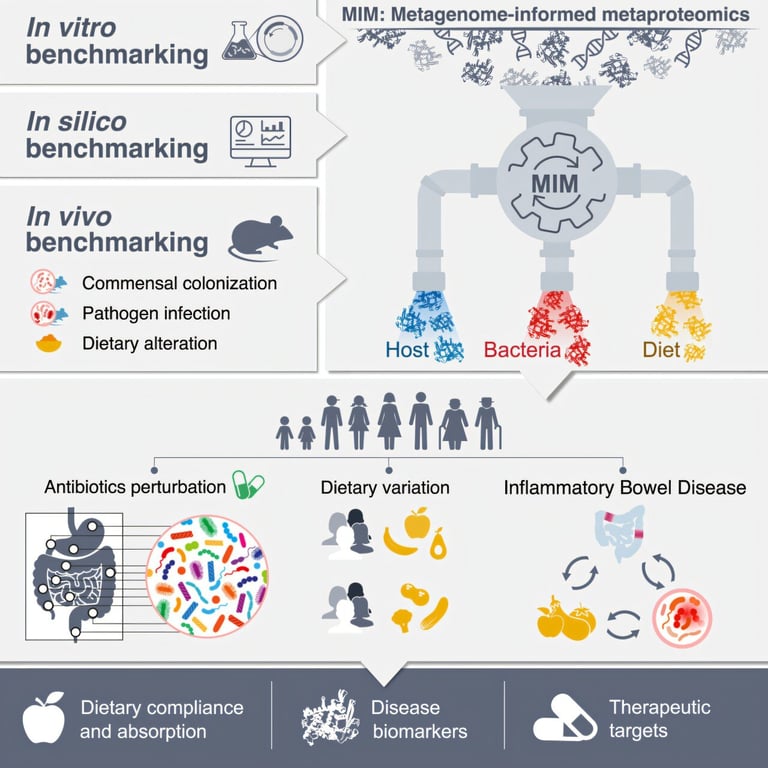
IPHOMED is capable of distinguishing proteins from various bacterial strains as well as those secreted by the human gut, creating a detailed atlas of gut-microbiome interactions.
Through this method, scientists identified previously unknown antimicrobial peptides in the human gut that play a crucial role in shaping individual microbiomes, thereby explaining variations in disease susceptibility.
The technique also tracked food consumption among volunteers, accurately identifying specific dietary items, such as peanuts, and monitoring adherence to diets in patients with gastrointestinal diseases.
Researchers at the Weizmann Institute of Science have unveiled a groundbreaking method for analyzing proteins in the intestine, which offers valuable insights into lifestyle and health.
This innovative technique, known as IPHOMED (Integrated Proteo-genomics of HOst, MicrobiomE and Diet), merges DNA sequencing with mass spectrometry to construct a personalized protein map.
In their studies, the researchers applied this method to inflammatory bowel disease, uncovering new proteins that could serve as potential drug targets and biomarkers for diagnosis and monitoring.
To facilitate this tracking, the researchers developed a comprehensive database of food proteins, allowing for unprecedented detail in assessing individual dietary intake through stool samples.
Published in the journal Cell, the study highlights how the method can identify all proteins present in stool samples, encompassing those derived from food, the human body, and the intestinal microbiome.
This breakthrough paves the way for enhanced treatment strategies for a range of disorders influenced by the microbiome, including inflammatory and neurodegenerative diseases.
Dr. Eran Elinav emphasized that identifying proteins is more crucial than DNA sequencing, as proteins provide insights into active bacterial functions and their impacts on human health.
The research team, led by Drs. Rafael Valdés-Mas, Avner Leshem, and Danping Zheng, collaborated with Dr. Alon Savidor, focusing on the microbiome's significant role in health.
Initially, the method successfully identified 97% of stool sample proteins, with the remaining 3% linked to dietary proteins, underscoring its potential applications in nutritional science.
Summary based on 1 source
Get a daily email with more Science stories
Source

Medical Xpress • Jan 20, 2025
We know what you ate: Detailed protein activity maps assess intestinal health