Revolutionary OncoIndx® Platform Achieves 98% Sensitivity in Cancer Genomic Profiling
October 7, 2024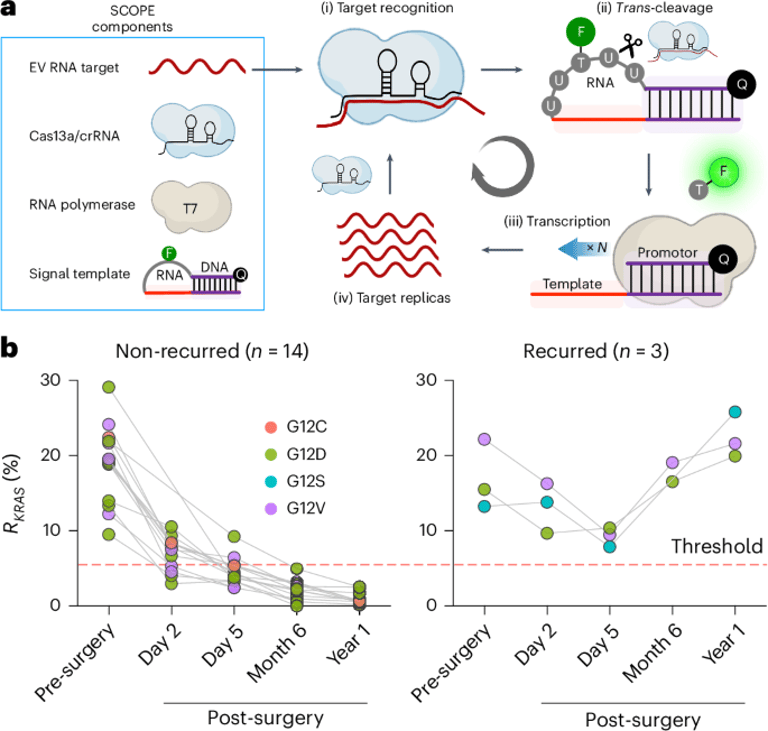
Additionally, the assay provides insights into tumor mutational burden (TMB), homologous recombination deficiency (HRD), and genomic loss of heterozygosity (gLOH), further enhancing its clinical relevance.
A novel CRISPR-based assay developed in the study achieves sub-attomolar sensitivity for recognizing and amplifying target mRNA, showcasing its potential for clinical applications.
A groundbreaking study published in Nature Biotechnology on October 7, 2024, by authors including Song, J. et al., introduces innovative techniques in genomic profiling and cancer diagnostics.
The research emphasizes the critical role of comprehensive genomic profiling (CGP) in precision oncology, advancing personalized medicine.
Central to this study is the OncoIndx® platform, a next-generation sequencing (NGS) assay designed to identify essential mutations that inform therapeutic decisions in cancer treatment.
OncoIndx® has demonstrated remarkable accuracy, achieving a maximum clinical sensitivity of 98% and a positive predictive value of 100% for actionable genomic variants.
The assay's high sensitivity and specificity could enhance liquid biopsy techniques, addressing significant implementation challenges in clinical settings.
The study also explores pooled CRISPR screens, which facilitate the parallel measurement of cellular responses to genetic changes using DNA-encoded barcodes.
A new technique, CRISPRmap, enhances the detection of cellular barcodes through optical readout, improving analysis in various cell types, including primary and stem cells.
Single-cell multimodal profiling, which examines both proteins and RNA, is highlighted as essential for a more accurate understanding of cellular functions.
The OncoIndx® assay targets major exons and selected introns of 1,080 cancer-associated genes, analyzing complex biomarkers such as single nucleotide variants (SNVs), copy number alterations (CNAs), and specific gene fusions.
Results indicate that CRISPRmap can accurately replicate known phenotypes related to specific base edits, offering valuable insights into gene functionality and cellular responses.
Summary based on 3 sources