New Study Decodes Chromatin Readers' Interactions with DNA
March 6, 2024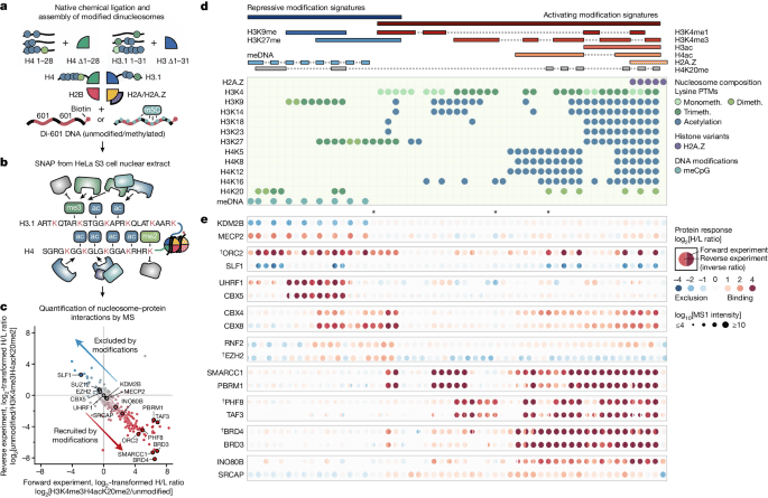
A recent study published in Nature introduces a new strategy using multidimensional mass spectrometry-based chromatin profiling to understand chromatin reader behavior.
Researchers created a library of nucleosomes with biologically relevant modifications to systematically profile over 1,900 proteins and their responses to different chromatin states.
The interactive online resource MARCS was developed, providing access to heat maps and data visualization tools to analyze chromatin reader interactions.
The study revealed two principal modes by which chromatin readers interact with modifications: simple responses to single modifications and complex patterns indicating responses to multiple modifications or synergistic effects.
The research provides insights into the complex network of protein behaviors in response to chromatin modifications, indicating that many proteins respond to multiple chromatin features.
Notable findings include the differential protein binding to specific chromatin marks such as H3K4 methylations, which have distinct roles in promoters and enhancers.
The study's data and tools like MARCS are expected to significantly advance future chromatin research and understanding of the epigenetic regulation.
Summary based on 1 source
Get a daily email with more Science stories
Source

Nature • Mar 6, 2024
Decoding chromatin states by proteomic profiling of nucleosome readers