Study Uncovers Unexpected Prevalence of Inverted D Segments in Antibody Diversity
January 29, 2025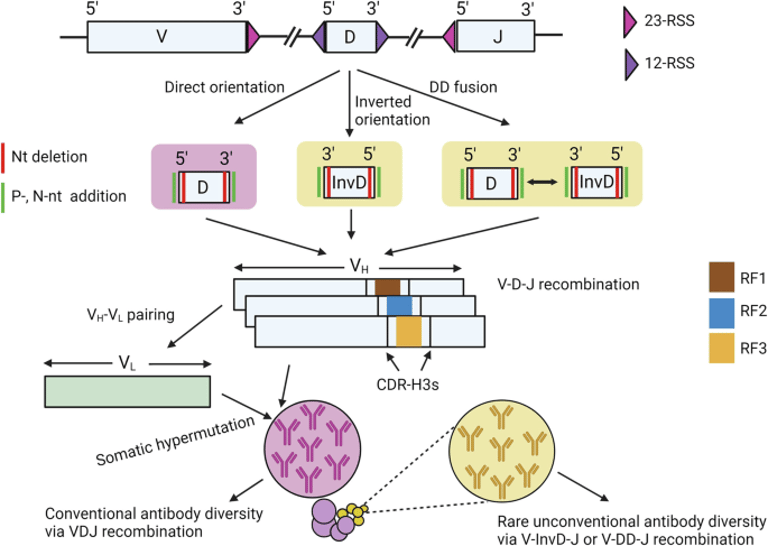
Recent research analyzed nearly 3 billion annotated VH sequences from 13 healthy individuals, revealing a greater prevalence of inverted D segments (InvDs) than previously recognized.
This study challenges prior beliefs about the rarity of InvDs and their limited role in antibody diversity, opening new avenues for therapeutic development and enhancing our understanding of immune responses.
D-D fusions, which can include InvDs, were found to significantly contribute to antibody diversity, with a notable increase in memory B cells.
The diversity of antibodies is critical for neutralizing various pathogens, arising from genetic recombination in B cells involving V, D, and J gene segments.
D genes significantly shape the complementarity-determining region 3 of the heavy chain (CDR-H3) through their recombination process.
InvDs can utilize all three reading frames, contributing to unique CDR-H3 diversity and enhancing antibody repertoire complexity.
D genes have identical 12 base pair spacers at both ends, suggesting a potential for bidirectional recombination, which may enhance antibody diversity.
Distinct amino acid motifs associated with InvDs, particularly histidine-rich and proline-rich stretches, were identified, which may influence antibody function.
Over two dozen human antibodies with InvDs were documented, demonstrating their role in targeting various antigens, including HIV and SARS-CoV.
Previous studies identified only a few inverted D segments (InvDs) and D-D fusions due to limitations in sequencing technologies and methods.
Summary based on 1 source